

| Python code for SVM |
|---|
| Cleaned data |
| Python Code to SVM |
SVM also called Support Vector Machine is a very strong tool when we apply it on classification and regression, and it will maximize the prediction accuracy of a model without overfitting the training data. SVM works by mapping data to a high-dimensional feature space so that data points can be categorized.
The objective of SVM algorithm is to find a hyperplane in an N-dimensional space that can distinctly classifies the data points. The dimension of the hyperplane depends upon the number of features. If the number of input features is two, then the hyperplane is a line. If the number of input features is three, then the hyperplane is a 2D plane. It becomes difficult to imagine when the number of features exceeds three.
There is also a very important concept in SVM which is kernels. Their job is to take data as input, and they transform it in any required form. We usually have: 1. Linear Kernel Function: This kernel is one-dimensional and is the most basic form of kernel in SVM. 2.Gaussian Kernel Radial Basis Function (RBF): RBF is the radial basis function. This is used when there is no prior knowledge about the data. 3.Sigmoid Kernel: This is mainly used in neural networks. And is equivalent to a two-layer, perceptron model of the neural network. 4.Polynomial Kernel: The polynomial kernel is a general representation of kernels with a degree of more than one. It's useful for image processing.
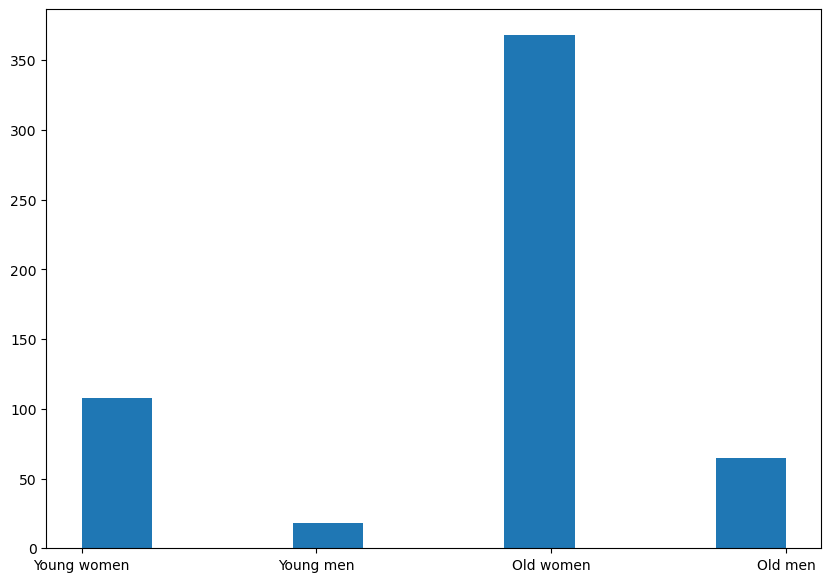
We can see this is an imbalanced data which may cause some class to be highly Correlated and the result will not be precise. I use smote to generate more sample in order to make it a balanced data. Where SMOTE is an oversampling technique where the synthetic samples are generated for the minority class and the advantages are it can overcome the overfitting problem posed by random oversampling.

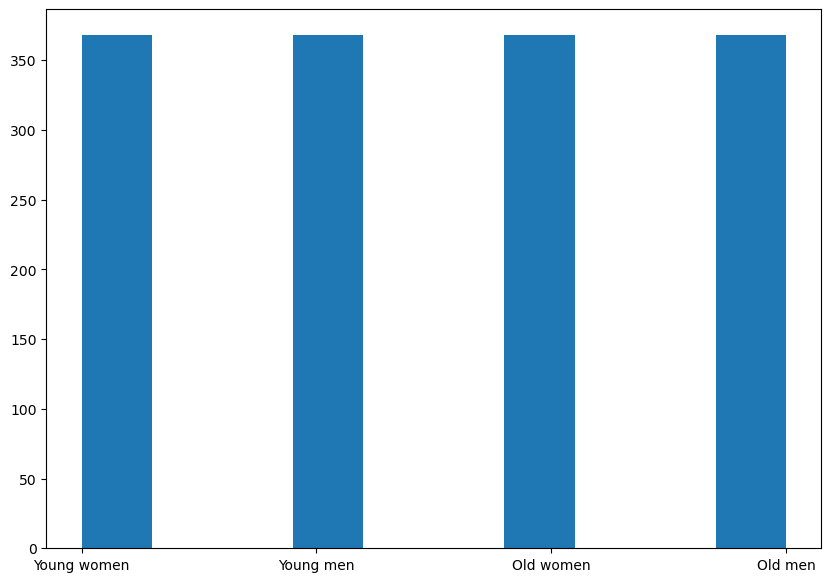
And this is distribution after making it become a balanced data, that all of the classes have the same number of samples.
I use 4 kernels linear, poly, rbf and sigmoid to see which kernels is the best for this model. From the 4 kernels' accuracy it shows that linear kernels are the most accurate one which is about 76.3%. And I also calculated the precision score, recall score and f-1 score. Since Precision is a good measure to determine, when the costs of False Positive are high. And Recall is a good model metric to determine, when there is a high cost associated with False Negative. F1 Score is the best when you want to seek a balance between Precision and Recall. And based on our problem about which how the drug AMITRIPTYLINE works, I will choose F1 score as the metric. And in the Linear kernels the young men group has the highest f1 score which is about 0.84.
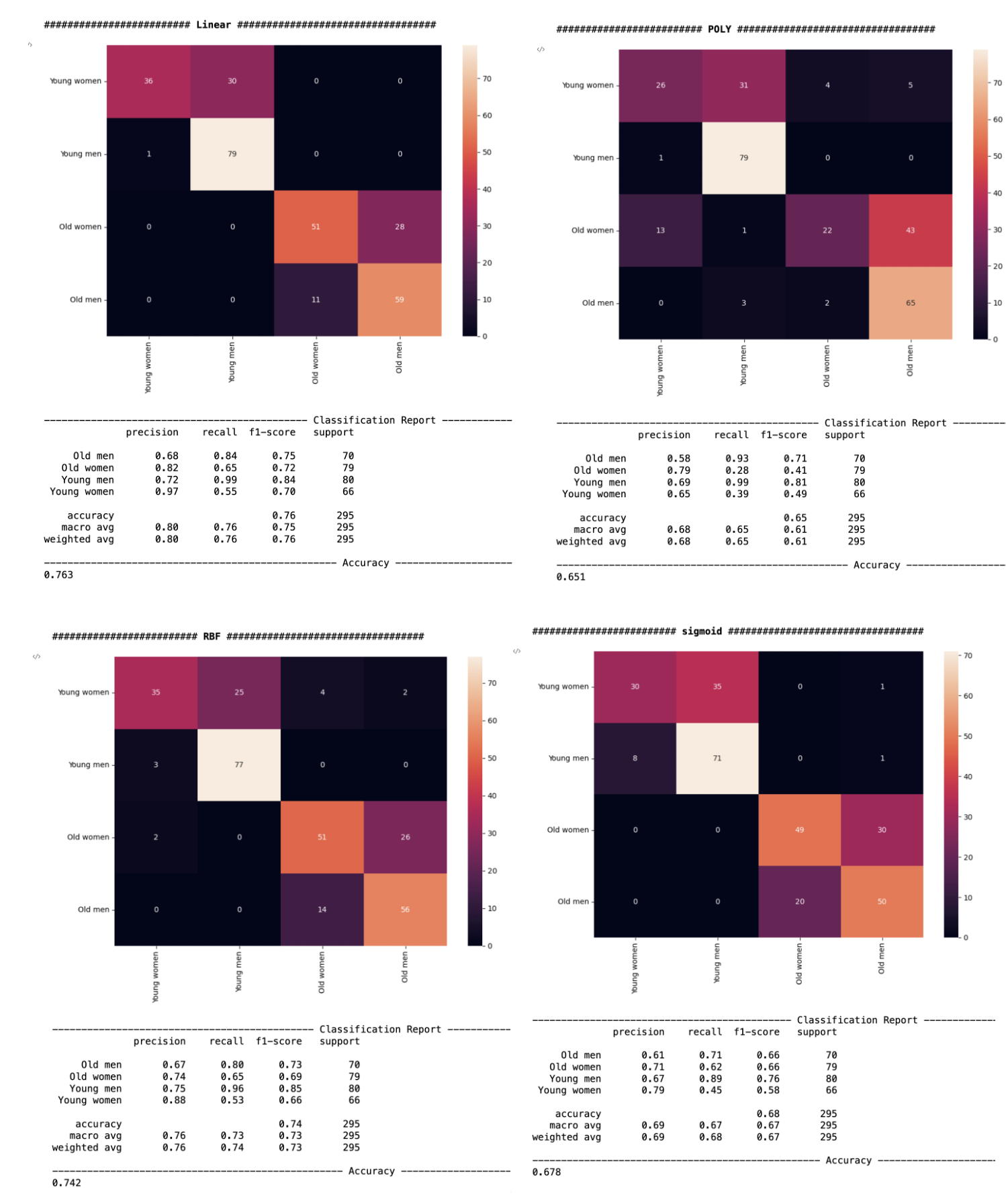
We will use Linear kernels to test the drug effectiveness since the accuracy is 76.3%. And poly kernels accuracy is 65.1%, RBF kernels accuracy is 74.2%, sigmoid kernels accuracy is 67.8%. The drug AMITRIPTYLINE was most helpful for the young male population within the group that took amitriptyline. And in the code section, we can find out the effectiveness of other drugs by simply changing the name of the drug.
Also, in the RBF kernels which the accuracy is about the same with Linear kernels, the young men group also have the highest f1 score. So, AMITRIPTYLINE is significant for depression treatment.